My laboratory has contributed to the area of malaria biology, particularly in molecular analyses of parasite plastid (apicoplast) function and field-based population genetics studies to understand malaria susceptibility.
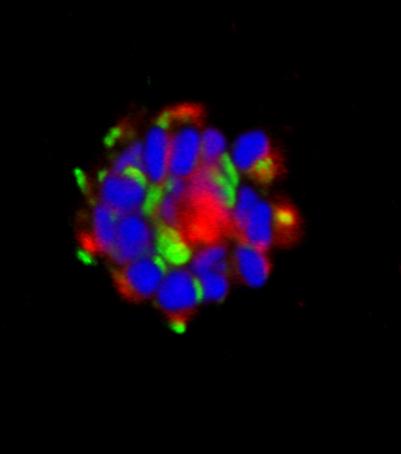
A P. falciparum schizont (apicoplasts in green)
The apicoplast is a four-membrane bound secondary endosymbiont that is essential for parasite survival, thus offering avenues for exploration of proteins and pathways for drug intervention against malaria. We have addressed the following:
Translation mechanisms of the Plasmodium apicoplast and rationale for the use of prokaryotic translation inhibitors against malaria.
The apicoplast genome encodes proteins rRNAs, tRNAs and ribosomal proteins in addition to three other ORFs, including one encoding the translation elongation factor EF-Tu. The absence of direct evidence for active translation in the apicoplast prompted us to investigate the phenomenon by generation of specific antibodies against apicoplast encoded EF-Tu and using these to investigate organellar localization and expression of the protein at different erythrocytic stages. This provided the first direct evidence for the apicoplast as a translationally active organelle (Mol. Microbiol., 2005). Under a project funded by the European Commission (EU-FP7), our group further investigated the binding of nuclear-encoded apicoplast-targeted EF-Ts with EF-Tu and demonstrated that interaction between these two translation factors, encoded by different cellular compartments, is a critical step in apicoplast protein synthesis (Int. J. Parasitol., 2011a). Our experiments showed that plastid-encoded EF-Tu possesses chaperone-related disulfide reductase activity that may have a role in rapid organellar response to protein misfolding or related stress in the apicoplast; this provides a rationale for the evolutionary retention of the tufA gene on the plastid genome. EF-G of the Plasmodium mitochondria and apicoplast was also explored as a possible drug target against malaria (TRENDS Parasitol., 2011). We established the role of two organelle targeted proteins as ribosome recycling factors of the parasite apicoplast and mitochondrion (Mol. Microbiol., 2013) and recently defined primary and ancillary functions of apicoplast and mitochondrial initiation factors in Plasmodium (Mol. Microbiol., 2015). Our work has important implications in the development of novel anti-malarials as well as the use of known antibiotics that target prokaryotic translation in anti-malarial chemotherapy (Mol. Biochem. Parasitol., 2013). Recent work from our laboratory has also explored the unique composition of the reduced ribosomes (Open Biol., 2014) as well as stop codon recognition in Plasmodium organelles (Mol. Microbiol., 2016, in press).
Evidence for iron-sulfur cluster biogenesis machinery unique to the Plasmodium apicoplast
Several important reactions in the apicoplast require assembled [Fe-S] prosthetic groups on participating proteins as well as the reductant activity of ferredoxin that is converted from its apo- form by the assembly of [Fe-S] clusters inside the apicoplast. The [Fe-S] assembly pathway involving SUF proteins had been predicted to function in the apicoplast with one component (PfSufB) encoded by the plastid genome itself. Our group provided the first experimental evidence for the existence of the apicoplast SUF pathway by demonstrating the enzymatic activity of recombinant P. falciparum nuclear-encoded SufC and showing its localization in the apicoplast. Further, an internal region of apicoplast SufB was used to detect PfSufB-PfSufC interaction in vitro which was also confirmed in vivo. As a departure from bacterial SufB and similar to reported plant plastid SufB, apicoplast SufB exhibited ATPase activity suggesting the evolution of specialised functions in the plastid counterparts. Unlike bacterial SUFs, the expression of PfSufB and PfSufC was not altered by oxidative stress indicating that the SUF system plays a housekeeping role in the apicoplast (Int. J. Parasitol. 2011b). These results have provided the first experimental evidence for an active SUF machinery in the Plasmodium apicoplast, a pathway that is absent in humans and thus offers a unique and hitherto unexplored site for drug intervention and discovery efforts. Importantly, the SUF pathway has now been shown to be essential for parasite survival and recent results from our laboratory have delineated the first step of the pathway, i.e. mobilization of sulphur from free cysteine that involves participation of two key enzymes-SufS and SufE. The possibility of inhibiting this pathway by chemical intervention has also been demonstrated by us recently (Antimicrob. Agents Chemother., 2014).
Mechanism of DNA replication and organization of the Plasmodium falciparum apicoplast genome
Although inhibition of replication of the circular apicoplast DNA genome (plDNA) by known antibiotics causes parasite death, the mechanism of the replication process was unknown. As a first step, our group identified replication initiation sites (ori) of plDNA (Mol. Biochem. Parasitol., 2003) and demonstrated that that plDNA replication initiates with differential efficiencies at multiple sites within the inverted rerpeat (IR) region of the molecule during the D-loop/bi-directional ori replication mode of replication (Mol. Biochem. Parasitol., 2005). Analyses of replication-ori interacting proteins resulted in the identification of a nucleus-encoded protein chaperone DnaJ homolog that interacts with ori elements in the apicoplast thus indicating its role in apicoplast DNA replication (Mol. Microbiol., 2010). The 35 kb apicoplast genome condenses into a small organelle prompting investigation of the protein(s) involved in DNA organization. This was explored through the histone-like (HU) protein that is the primary organizational component of bacterial nucleoids, dinoflagellate chromosomes as well as red algal chloroplast genomes. Nuclear-encoded PfHU was shown to be expressed throughout the intra-erythrocytic phase of parasite growth and localized within the apicoplast. PfHU also interacted specifically with plDNA (as opposed to nuclear DNA). It exhibited DNA binding and was capable of condensing DNA into bundles and bridges. PfHU was incapable of introducing negative supercoils in relaxed DNA but induced linear DNA concatenation in a concentration dependent manner. This established PfHU as the primary DNA condensation protein of the apicoplast genome and highlighted its significance in the process of apicoplast DNA replication and organelle division (Nucleic Acids Res., 2008).
Analysis of genetic factors, particularly polymorphisms in human host molecules involved in immune regulation, adhesion, rosetting and invasion of the malaria parasite in susceptibility/resistance to falciparum malaria in India.
The contribution of malaria as a selection force for specific human genetic polymorphisms is well known in world populations. However, there was absence of information on genetic variations in Indian populations and their association with malaria susceptibility/resistance to falciparum malaria in India. This is an important issue as P. falciparum-endemic regions of India, such as parts of Orissa, are home to a large number of tribal populations that represent a unique genetic pool among Indian population groups (IGVC, J. Genet., 2008a). Our laboratory initiated work on understanding the role of human genetic variation in susceptibility to falciparum malaria in India, first as part of the Indian Genome Variation Consortium (IGVC) (Hum. Genet., 2005; J. Genet., 2008a) and subsequently with funding from the DBT.
We set-up a collaboration with clinicians and field researchers from KGMU (Lucknow), NIMR (New Delhi), and IGH (Rourkela), to gather detailed clinical and genetic information from patients and controls from Uttar Pradesh and the primarily tribal belt of Orissa and Chattisgarh. This study has provided interesting information on the significance of genetic variation in immune regulatory molecules (TNF and FcγRIIa) vis-a-vis their levels in patients and association with severe disease manifestation for TNF and with protection from disease for the high IgG2-binding FcγRIIa His/His (A/A) genotype (Malar. J., 2008a). Analysis of variations in host adhesion molecules that play a significant role in the pathogenesis of falciparum malaria also revealed the significance of variations in the ICAM1, PECAM1 and CD36 genes in the manifestation of disease in India (Malar. J., 2008b). Complement receptor CR1/CD35 on erythrocytes is involved in rosetting (a phenomenon associated with severe manifestation of malaria) and merozoite invasion. Our study identified two CR1 SNPs that define the low expression (L) CR1 allele in Indian populations and showed that populations of the malaria endemic region have very low RBC surface CR1 levels and higher frequencies of the L alleles with high CR1 levels associated with manifestation of disease (Hum. Immunol., 2009). The immune effector response to P. falciparum infection and the processes by which they mediate the development of clinical immunity in areas with different endemicity are poorly understood. Our analysis of circulating levels of pro-inflammatory and anti-inflammatory cytokines and the relative balance between the Th1 and Th2 response illustrated how populations residing in areas of varying disease endemicity may respond to P. falciparum-induced immune challenge (Eur. Cyt. Netw., 2010). Work in collaboration with CSIR-IGIB established the significant association of the human APOBEC3B gene deletion with susceptibility to falciparum malaria in India (Infect. Genet. Evol., 2012) and collaborative work with Oxford University, UK helped understand negative epistasis between α+ thalassaemia and sickle cell trait as an explanation for inter-population variation in India (Evolution, 2011). Recent work from our laboratory showed that an intron 3 and a 3’UTR SNP of IFNG associated with disease in the endemic region. In addition, large (CA)n repeats of IFNG intron1 correlated with protection from disease manifestation with a stronger association observed for protection from severe malaria in the endemic region (Infect. Genet. Evol., 2015). Differential contribution of SNPs and microsatellite repeats of adhesion molecules THBS1 and ESEL, and immune regulatory molecule genes NOSII, CRP was also observed (Eur. J. Clin. Microbiol. Infect. Dis., 2015).