Viral diseases account for a major proportion of human infectious diseases in India. Many of them are caused by RNA viruses and follow zoonotic, arthropod-borne, or aerosol routes of transmission. One such virus, SARS CoV2, responsible for COVID 19 pandemic, is a grim reminder of the lack of preparedness in viral diagnostics, curtailing viral transmission, and interventional strategies to manage the disease. At the same time, however, this pandemic has set a benchmark for the development, approval, and marketing of novel prophylactic, diagnostic and therapeutic technologies in the shortest time. Numerous prevalent viral infections, either due to the worldwide presence of intermediate host or the emergence of genetic variants with altered transmission patterns, pose a risk of outbreaks in new locations and global spread. Remaining pandemic ready is a priority going forward for the safety of public health in India. In our lab, we are trying to address these questions using a multitude of approaches based on our expertise in virology, molecular biology, and cell biology.
1. Rapid vaccine platforms for emerging viral diseases.
Vaccine platforms based on adenovirus, VLP, subunit, or mRNA vaccines can be customized and developed in a short duration. Our goal is to create rapid and affordable vaccine platforms against viruses of the epidemic potential in India. We are focused to develop and evaluate adenovirus and VLP-based vaccine candidates against viruses of epidemic potential.
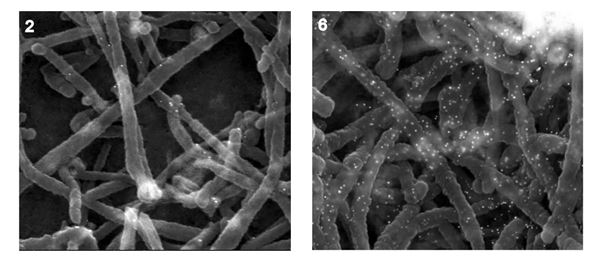 Fig :
Fig : High-resolution analysis of VLPs generated by transfecting select respiratory syncytial virus (RSV) proteins (6) compared to RSV particles on infected cells (2). Meshram et al 2016 (JVI).
2. Development of diagnostics for the detection of viral diseases.
Early detection of viruses plays an important role in developing strategies to curtail transmission in case of a widespread outbreak. In our lab, we seek to develop molecular tests such as PCR or RT-PCR or a rapid and simple loop-mediated isothermal amplification (LAMP) for the detection of emerging and re-emerging viruses in clinical samples.
3. Tracking new variants of virus and immunity status in the population.
The uncertain possibility of the next viral pandemic requires us to track the emergence and transmission of new strains of prevalent as well as emerging viruses. We aim to use cutting-edge diagnostic techniques for the detection of viruses in clinical samples followed by sequencing to confirm the emergence of variants. The second objective is to monitor current vaccine-induced immunity against new strains of viruses (particularly respiratory viruses) circulating in the human population. We will use a virus-based serum neutralization assay to monitor the protectiveness of vaccine-induced immunity in the human population. This strategy is particularly important to plan changes in vaccine formulations for better immunoprotection.
4. Assay development for antiviral drug screening.
The development of assays to evaluate the antiviral effects of hit compounds targeting virus entry and fusion, genome replication and transcription rate, assembly, and the infection rate is crucial in antiviral drug development. Our lab is strived to develop in vitro assays for drug screening against important viruses such as coronaviruses, JE, influenza, chikungunya, Dengue virus, and many others. We plan to develop assays based on the following approaches: 1. Plaque forming assay or TCID50 to test antiviral effects of compounds. 2. Virus-like particles (VLP) or pseudovirus-based assays to screen compounds that target viral fusion, entry, and fusion events. 3. Self-replicating subgenomic viral RNA (replicon)-based assay to evaluate drugs that target genome replication and transcription. The last two approaches are mainly designed to evaluate drugs against BSL3 and BSL4 viral agents at the BSL2 level obviating the need to handle highly pathogenic viruses. For preclinical evaluation of lead molecules, in vivo infectious disease animal models will be developed at BSL2 and BSL3 levels.
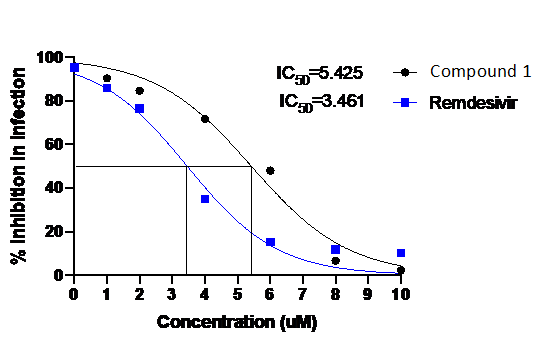 Fig :
Fig : Dose-response curve of compound1 compared to Remdesivir against SARS CoV2 (MOI-0.1) in Vero cells. Unpublished.
5. Host-viral interaction studies for target identification in drug development studies.
To catalyze antiviral drug discovery, it is vital to understand molecular virus-host interactions happening during viral infection. Specific viral proteins must interact with host proteins during viral genome replication, virus assembly, and release. Disruption of these interactions is detrimental to virus multiplication. Identification and validation of host or viral targets critical in virus replication provide a jump start in designing novel antiviral drug molecules for therapeutics development against viruses. In our lab, we plan to identify and validate the viral and/or host proteins that will serve as antiviral targets. We will use the classical affinity purification-mass spectrometry (AP-MS) approach to construct the virus-human protein-protein interaction (PPI) network and followed by an insilico molecular docking approach to identify and redesign potential drugs.
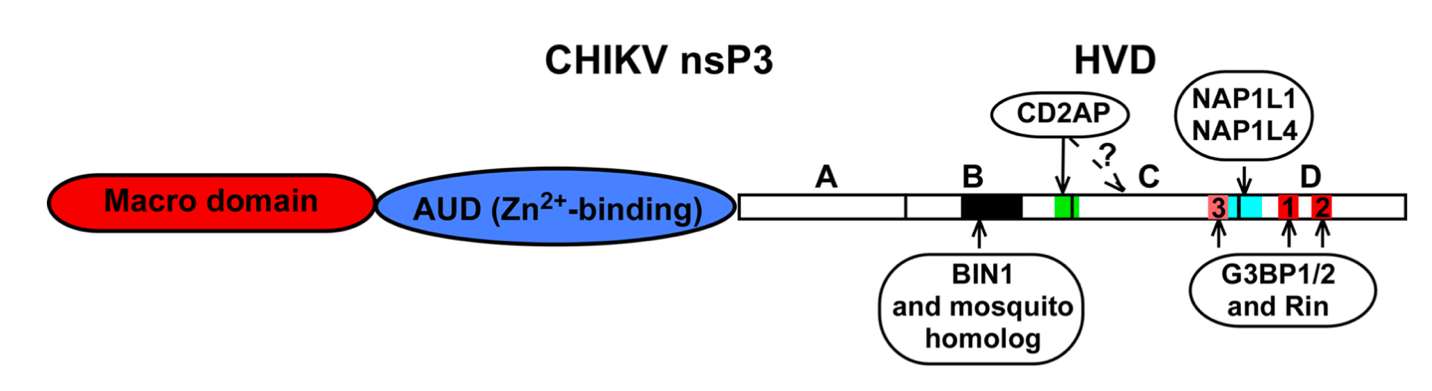 Fig :
Fig : Binding sites of cellular proteins on the hypervariable domain (HVD) of nonstructural protein (nsP3) of chikungunya virus (CHIKV) that play a critical role in virus replication. Meshram et al 2018 (JVI).
6. Reverse genetics system (RGS) for RNA viruses.
Infectious cDNA clone of RNA viruses is an essential tool to study the function of viral genes in viral processes such as replication, transcription, assembly, and pathogenesis. Using our previous expertise to develop RGS in respiratory syncytial virus (RSV) and alphaviruses, we plan to develop a reverse genetic system for emerging RNA viruses in India that will explore molecular events during the infectious life cycle of viruses and help develop viral vaccine candidates.
About Dr. Meshram: He is a Senior Scientist in the Division of Virus Research and Therapeutics. He has 14 years of experience in studying zoonotic, mosquito-borne, and respiratory viruses. His major expertise is in the generation of reverse genetics of RNA viruses, host-viral interaction to identify targets for therapeutics development, development of animal models for infectious disease, antiviral drug screening, development and evaluation of vaccine candidates, and development of diagnostic tools.
Expertise: Virology, Molecular biology, Vaccine, Diagnostics, BSL3 agents, and RNA viruses